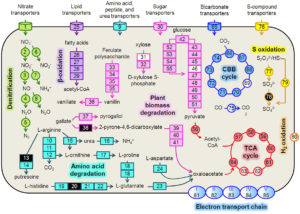
Summary of key metabolic pathways expressed by a prominent bacterium (Hydrogenophaga b174) in an NRZ biogeochemical hot spot in the Rifle aquifer. Surprisingly, this bacterium actively catalyzed both heterotrophic and chemolithoautotrophic processes and influenced biogeochemical cycling of several elements, including C, N, and S. Unexpectedly, denitrification played an important role in this metabolism.
Organic matter deposits in alluvial aquifers have been shown to result in the formation of NRZs, which can modulate aquifer redox status and influence the speciation and mobility of metals, significantly affecting groundwater geochemistry. This study (Jewell et al., Frontiers in Microbiology) sought to better understand how natural organic matter fuels microbial communities within anoxic biogeochemical hot spots (NRZs) in a shallow alluvial aquifer at the Rifle (CO) site. Overall, the results highlighted the complex nature of organic matter transformation in NRZs and the microbial metabolic pathways that interact to mediate redox status and elemental cycling.
Summary
The authors used an anaerobic microcosm experiment in which NRZ sediments served as the sole source of electron donors and microorganisms. Biogeochemical data indicated that the decomposition of native organic matter occurred in different phases, beginning with mineralization of dissolved organic matter (DOM) to CO2 during the first week of incubation, followed by a pulse of acetogenesis that dominated carbon flux after two weeks. The depletion of DOM over time was strongly correlated with increases in expression of many genes associated with heterotrophy (e.g., amino acid, fatty acid, and carbohydrate metabolism) belonging to a Hydrogenophaga strain that accounted for a relatively large percentage (~8%) of the metatranscriptome. This Hydrogenophaga strain also expressed genes indicative of chemolithoautotrophy, including CO2 fixation, H2 oxidation, S-compound oxidation, and denitrification. The pulse of acetogenesis appears to have been collectively catalyzed by a number of different organisms and metabolisms, most prominently pyruvate:ferredoxin oxidoreductase. Unexpected genes were identified among the most highly expressed (>98th percentile) transcripts, including acetone carboxylase and cell-wall-associated hydrolases with unknown substrates. Many of the most highly expressed hydrolases belonged to a Ca. Bathyarchaeota strain and may have been associated with recycling of bacterial biomass.