Sulfate is ubiquitous in the environment, and sulfate reduction – a key control on anaerobic carbon turnover – impacts a number of other processes such as carbon oxidation and sulfide production. Until now, sulfate reduction was believed to be restricted to organisms from select bacterial and archael phyla. But scientists at UC Berkeley have now found this ability to be more widespread. They used genome-resolved metagenomics to discover roles in sulfur cycling for organisms from 16 microbial phyla not previously associated with this process.
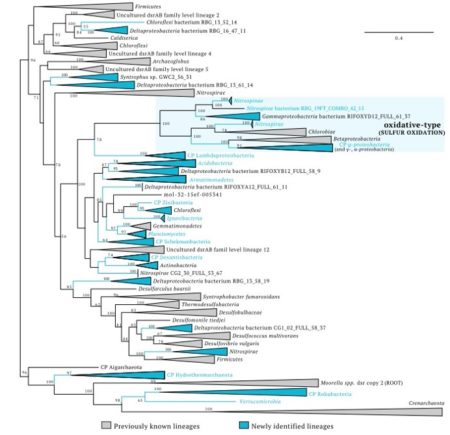
DsrAB protein tree showing the diversity of organisms involved in dissimilatory sulfur cycling using the dsr system.
Lineages in blue contain genomes reported in this study. Phylum-level lineages with first report of evidence for sulfur cycling are indicated by blue letters.
Sulfate-reducing bacteria are anaerobic microorganisms essential to sulfur and carbon cycling. Sulfate reduction drives other key processes and produces hydrogen sulfide, an important but potentially toxic gas present in sediments, wetlands, aquifers, the human gut, and the deep-sea. The discovery of novel microbes connected to sulfur cycling is relevant in biogeochemistry, ecosystem science and engineering, and fundamentally reshape our understanding of microbial function and capabilities associated with phylogenetic information.
Summary
Phylogenetic information shapes our expectations regarding microbial capabilities. In fact, this is the basis of currently used methods that link gene surveys to metabolic predictions of community function. Sulfate Reduction, an important anaerobic metabolism, impacts carbon, nitrogen, and hydrogen transformations in numerous environments across our planet and is known to be restricted to organisms from selected bacterial and archaeal phyla. The authors used genome-resolved metagenomic analyses to determine the metabolic potential of microorganisms from six complex marine and terrestrial environments. By analyzing >4000 genomes, they identified 123 near-complete genomes that encode dissimilatory sulfite reductases involved in sulfate reduction. They discovered roles in sulfur cycling for organisms from 16 microbial phyla not previously known to be associated with this process. Additional findings include some of the earliest-evolved sulfite reductases in bacteria, identification of a novel protein unique to sulfate reducing bacteria, and a key sulfite reductase gene in putatively symbiotic Candidate Phyla Radiation (CPR) bacteria. This study fundamentally reshapes expectations regarding the roles of a remarkable diversity of organisms in the biogeochemical cycle of sulfur.
Citation
Anantharaman, K., B. Hausmann, S.P. Jungbluth, R.S. Kantor, A. Lavy, L.A. Warren, M.S. Rappé, M. Pester, A. Loy, B.C. Thomas, and J.F. Banfield (2017). Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. The ISME journal 12, 1715–1728, DOI: 10.1038/s41396-018-0078-0