Scientific Achievement
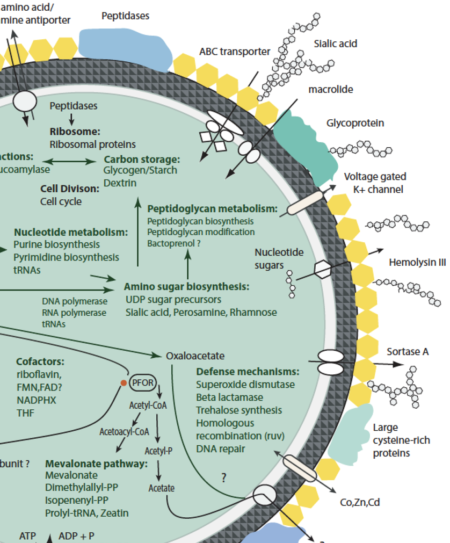
Complete genomes reconstructed for five closely related and highly novel groundwater-associated bacteria were used to predict their environmental impact. The genomes define a new Class in Peregrinibacteria, potentially a new phylum.
Significance and Impact
Complete genomes from complex metagenomes are essentially unprecedented. Having such high quality sequences is critical for accurate ecological and evolutionary analyses.
Research Details
- Field samples were collected, DNA extracted and shotgun sequenced
- Advanced bioinformatics approaches were used to recover draft genomes and curate them to completion
- Phylogenetic analyses place the newly sampled organisms in the Candidate Phyla Radiation as a sibling lineage to Peregrinibacteria.
Citation
Anantharaman, K.; Brown, C. T.; Burstein, D.; Castelle, C. J.; Probst, A. J.; Thomas, B. C.; Williams, K. H.; Banfield, J. F. (2016), Analysis of five complete genome sequences for members of the class Peribacteria in the recently recognized Peregrinibacteria bacterial phylum, PeerJ, 4, e1607, DOI: 10.7717/peerj.1607