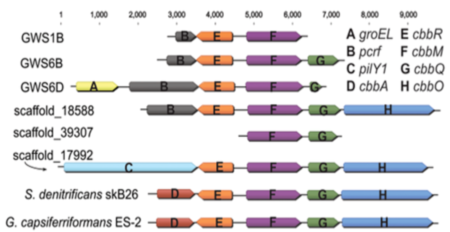
Operon structure alignments of the recovered metagenomic fragments from the Rifle groundwater aquifer, along with closest RubisCO hits and related sequences in NCBI. pcrf stands for peptide chain release factor.
Scientific Achievement
We report a functional metagenomic selection that recovers physiologically active RuBisCO molecules directly from uncultivated and largely unknown members of natural microbial communities.
Significance and Impact
Functional metagenomic selection leverages natural biological diversity and billions of years of evolution to provide a new window into the discovery of CO2‐fixing enzymes
Research Details
- Selection based on CO2‐dependent growth in a strain expressing environmental DNA
- RuBisCOs selected, purified and tested for CO2/O2specificity
- X‐ray crystallography: one enzyme is a hexamer, the second form II multimer ever solved and the first structure from an uncultivated bacterium.
Citation
Varaljay, V. A.; Satagopan, S.; North, J. A.; Witte, B.; Dourado, M. N.; Anantharaman, K.; Arbing, M. A.; McCann, S. H.; Oremland, R. S.; Banfield, J. F.; Wrighton, K. C.; Tabita, F. R. (2016) Functional metagenomic selection of ribulose 1,5-bisphosphate carboxylase/oxygenase from uncultivated bacteria, Environmental Microbiology, 18, 1187-1199 DOI: 10.1111/1462-2920.13138.