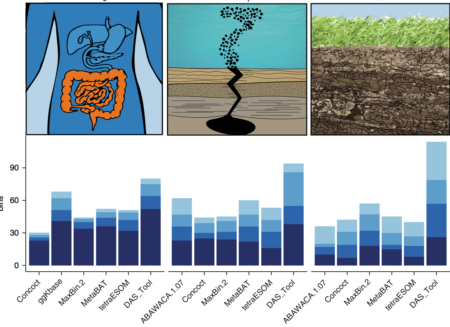
The number of high-quality genomes with low contamination from three ecosystems of varying complexity. Bin completeness increases with increasing shade of blue.
Genomes reconstructed directly from DNA sequences sampled from natural environments have revolutionized scientific understanding of microbial diversity and evolution. While this process can be difficult, a new automated method called DAS Tool integrates a flexible number of binning algorithms to calculate an optimized, non-redundant set of bins from a single assembly, thereby greatly improving the recovery of genomes from natural environments.
The recovery of genomes, especially from complex environments such as soil, will be facilitated by the new automated DAS Tool.
Summary
Understanding of the metabolic capacities of microorganisms in natural environments is critical to prediction of ecosystem function. Analysis of organism-specific metabolic pathways and reconstruction of community interaction networks requires high-quality genomes. However, existing binning methods often fail to reconstruct a reasonable number of genomes and report many bins of low quality and completeness. Furthermore, the performance of existing algorithms varies between samples and environment types. A dereplication, aggregation and scoring strategy, DAS Tool, was developed. This algorithm combines the strengths of a flexible set of established binning algorithms. DAS Tool applied to a constructed community generated more accurate bins than any automated method. Indeed, when applied to environmental and host-associated samples of different complexity, DAS Tool recovered substantially more near-complete genomes, including those for organisms from previously unreported lineages, than any single binning method alone. The ability to reconstruct many near-complete genomes from metagenomics data will greatly advance genome-centric analyses of ecosystems.
Citation
C.M.K. Sieber, A.J. Probst, A. Sharrar, B.C. Thomas, M. Hess, S.G. Tringe, and J.F. Banfield “Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy”, Nature Microbiology, 3, 836 (2018) DOI: 10.1038/s41564-018-0171-1