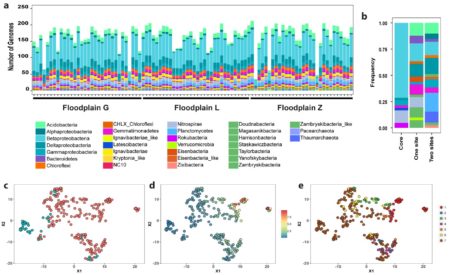
Taxa detected across samples (a). A core floodplain microbiome includes abundant Betaproteobacteria (b), and 42 genomes (c; teal) present in > 89 samples with a low coefficient of variation of abundance (d), not associated with any given floodplain (e; brown).
We studied the most abundant microorganisms (microbiome) in 130 topsoil samples from three meander-bound floodplains along the East River, CO during a period of low river flow and over two consecutive years. We reconstructed 248 draft quality genomes (at the sub-species level) from these samples. DNA (metagenomes) and RNA (metatranscriptome) sequences revealed the presence of bacteria that are commonly found across the floodplains over time. Despite the very high microbial diversity and complexity of the soils, ~15% of species were detected in two consecutive years, and approximately one third of the representative genomes were detected with similar levels of abundance across all three locations. The capacities for aerobic respiration, aerobic CO oxidation (and other small molecules), and thiosulfate oxidation were enriched in these microorganisms. However, the most active genes at the time of sampling were involved in nitrification, methanol and formate oxidation, and nitrogen and CO2 fixation. Our results highlight the prominence of sulfur, nitrogen, and one-carbon metabolism in the watershed.
We were able to reconstruct hundreds of microbial genomes from complex soil samples. We found that soils bounded by individual river meanders capture processes that occur in soils bounded by other meanders along the river corridor. This is important, given that there is a need to understand microbially-mediated biogeochemical transformations, which occur at the millionth of a meter scale, at the tens to hundreds of kilometer scale of watershed ecosystems. The presence of the same bacteria (~ 15%) over two consecutive years, combined with differences between common capacities and important capacities at the time of sampling, suggests that the floodplain soil microbiome is versatile and can respond to natural disturbances (e.g., flooding resulting from spring snowmelt).
Summary
Meander-bound floodplains appear to serve as scaling motifs that predict aggregate capacities for biogeochemical transformations in floodplain soils. We identified a core floodplain microbiome that was consistent across floodplains and that was enriched in capacities for aerobic respiration, aerobic CO (and other small molecules) oxidation, and thiosulfate oxidation with the formation of elemental sulfur. Systematic patterns of gene abundance based on sampling position relative to the river were not detected. The most highly transcribed genes in the middle floodplain were amoCAB and nxrAB (for nitrification) followed by genes involved in methanol and formate oxidation, and nitrogen and CO2 fixation. Additionally, low soil organic carbon correlated with high activity of genes involved in methanol, formate, sulfide, hydrogen, and ammonia oxidation, nitrite oxidoreduction, and nitrate and nitrite reduction. While widely represented genetic capacities did not predict in situ activity at one time point, they defined a reservoir of biogeochemical potential available as conditions change and suggests the value of meanders as a scaling motif to improve prediction of watershed biogeochemistry.
Citation
Matheus Carnevali, P.B., Lavy, A., Thomas, A.D. et al. Meanders as a scaling motif for understanding of floodplain soil microbiome and biogeochemical potential at the watershed scale. Microbiome 9, 121 (2021). https://doi.org/10.1186/s40168-020-00957-z