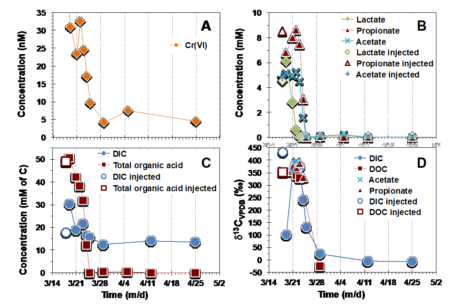
Charts showing the variations versus time of Cr (VI), electron donors, and metabolic products, in groundwater. The gray bars indicate the injection day of the electron donor. (A) Cr (VI) concentration over time; (B) organic acid concentration; (C) dissolved inorganic carbon (DIC) and total organic acids expressed as mM carbon; (D) δ13C values of organic acids and DIC.
Soils and groundwater contamination by hexavalent chromium Cr(VI) is common in industrial areas and is a serious threat to water quality and human health. In a field-scale experiment of microbial Cr(VI) reduction, a team of scientists used stable isotopes of carbon to demonstrate the transfer of carbon from the original electron donor (i.e., bacterial food source) to the metabolic products, and subsequent reduction of Cr(VI).
By injecting 13C-labeled electron donors into a contaminated site, scientists not only demonstrated that the existing microbial community reduced metal contaminants, but that this approach is a viable method for estimating the efficiency of biostimulation. This approach could be transferred to other contaminated sites that contain a variety of metal and organic contaminants.
Summary
Hexavalent chromium Cr(VI) is a common inorganic contaminant in soils and groundwater of industrial areas and represents a serious threat to water quality and human health. Among the various techniques currently available, in situ biostimulation has been recognized as a relatively cost-effective and valuable method for the remediation of contaminated groundwater. To date, the transformation and fate of organic electron donors used to stimulate Cr(VI) reduction in the field has been reported only in limited studies due to analytical and technical challenges. In this work, the authors report field-scale experimental results from in situ microbial Cr(VI) reduction stimulated via injection of 13C-labelled lactate. Simultaneously with Cr(VI) reduction the authors used concentrations and carbon isotope ratios of metabolic products to monitor the carbon transfer from the original 13C-labelled lactate. The authors also monitored the carbon isotope ratios of phospholipid fatty acids (PLFA) to demonstrate the transfer of carbon from 13C-labelled lactate to a portion of the microbial community.
Citation
M. Bill, M.E. Conrad, B. Faybishenko, J.T. Larsen, J.T. Geller, S.E. Borglin, and H.R. Beller, “Use of carbon stable isotopes to monitor biostimulation and electron donor fate in chromium-contaminated groundwater.” Chemosphere, 235, 440-446 (2019), DOI: 10.1016/j.chemosphere.2019.06.056